RESEARCH PROJECTS
During brain development, progenitor cells balance proliferation and differentiation to generate the enormous diversity of neurons in the correct numbers and proportions. We study the embryonic brainstem development in zebrafish to address some of the main unsolved questions in developmental neurobiology and regenerative research: How does the brain generate its enormous diversity of neurons in the correct numbers and proportions? What are the mechanisms behind this tightly coordinated and dramatic transformation that the brain undergoes —from the simple tubular structure of the neural tube, to the highly convoluted structure of the brain? Does the brainstem harbor cell populations that display different stemness properties over time? And how do genetic networks ultimately build and control complex organs, such as the brain? Zebrafish embryonic brainstem —the hindbrain— development is a good model to address these questions, as it requires both dynamic self-organization and dramatic morphogenetic changes for its final functional outcome. Further, and importantly, the hindbrain is a highly conserved brain structure in vertebrates.
We use the zebrafish embryo as a model system, which allows us to combine cutting-edge complementary approaches such as: CRISPR-Cas9 genome editing, high-resolution 4D-imaging paired with cell tracking tools, and analyses of gene regulatory landscapes. We would like to map the ontogeny of differentiated neurons, and to reveal the sequence of temporal transitions from progenitor to the differentiated state.
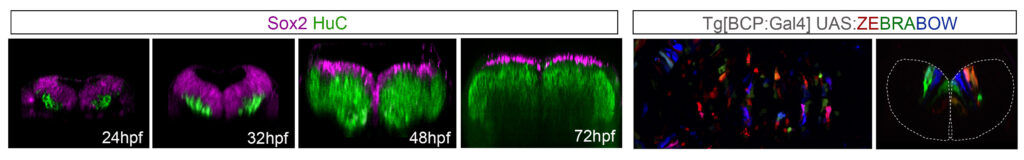
Reconstructing Cell Lineages
The genetic requirements for neurogenesis have been addressed, and the importance of bHLH factors has been revealed. Nevertheless, information about the relative spatiotemporal distribution of neural progenitor pools upon morphogenesis and their clonal relationships remains elusive due to the demands associated with the in vivo imaging of the embryonic brain. Therefore, reconstructing unambiguously the cell lineages is central to the understanding how the precise diversity of cell types develops and we need to incorporate time as a crucial but poorly understood factor.
To mirror reconstruction of the diversity of cell lineages we use high resolution 4D-imaging. We merge information from morphogenesis and cell fate gained by the use of complementary strategies: those using population average measures that provide information on proliferation kinetics and cell lineage specification, and those collecting information at the clonal level providing insight into the behavior of individual cells. Further, 4D-imaging and cell tracking can meet the challenge of unambiguous determination of single lineages, since the behavior of cells can be precisely monitored, and the lineages can be unequivocally reconstructed. We think that the combination of 4D-imaging and functional tools is crucial for closing the gap between genetics and cell biology.
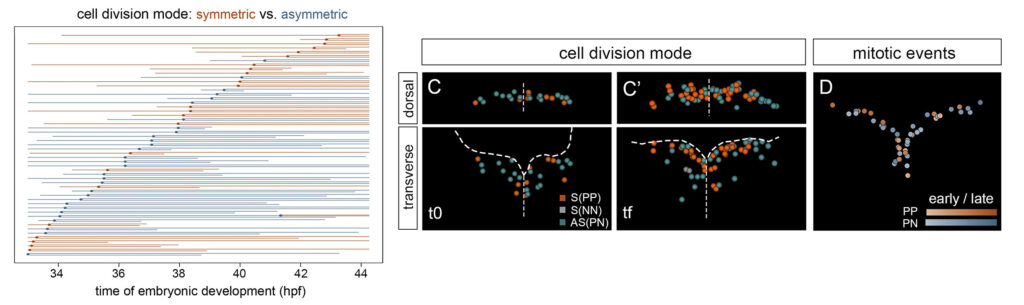
Studying tissue growth
The hindbrain undergoes dynamic self-organization with dramatic morphogenetic changes, which are crucial for the final functional outcome. To comprehend how the robust size of the hindbrain is accomplished with the proper cell composition, we study how multipotent progenitors transition towards different states by combining multicolor cell lineage tracing and functional tools.
We seek the contribution of the distinct progenitor cell populations to the whole differentiated neuronal domain, and to specific functional neuronal populations during hindbrain morphogenesis. We use genetic intersectional fate mapping, high-resolution in vivo imaging and the 4D-atlas of temporal differentiation.
We are interested in deciphering the cell state transitions that progenitor cell populations undergo to finally contribute to the neuronal circuits. For this, we investigate whether changes in cell progenitor states can be predicted by specific transcriptional signatures over time. Thus, we are dissecting the gene regulatory network operating in the generation of specific mature neuronal populations.
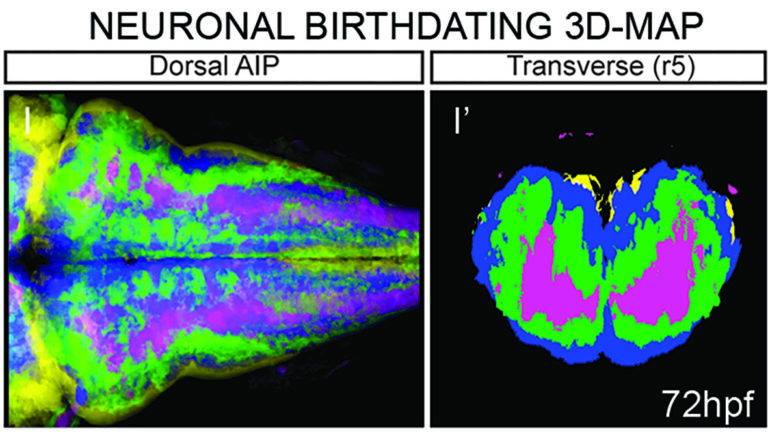
Building digital brain 3D-Atlas
We are developing computationally efficient and robust methods to build a digital representation of the developing vertebrate brain from 3D+time imaging data for further quantitative analysis and multilevel modeling. We are establishing a user-friendly pipeline for the brain 4D-atlas maker, which will allow to integrate high-throughput imaging data from different kind of experiments.
We have already generated digital neuronal birthdating 3D-maps and revealed that the temporal order of neuronal differentiation prefigured the spatial distribution of neurons in the tissue, with an inner-outer differentiation gradient.